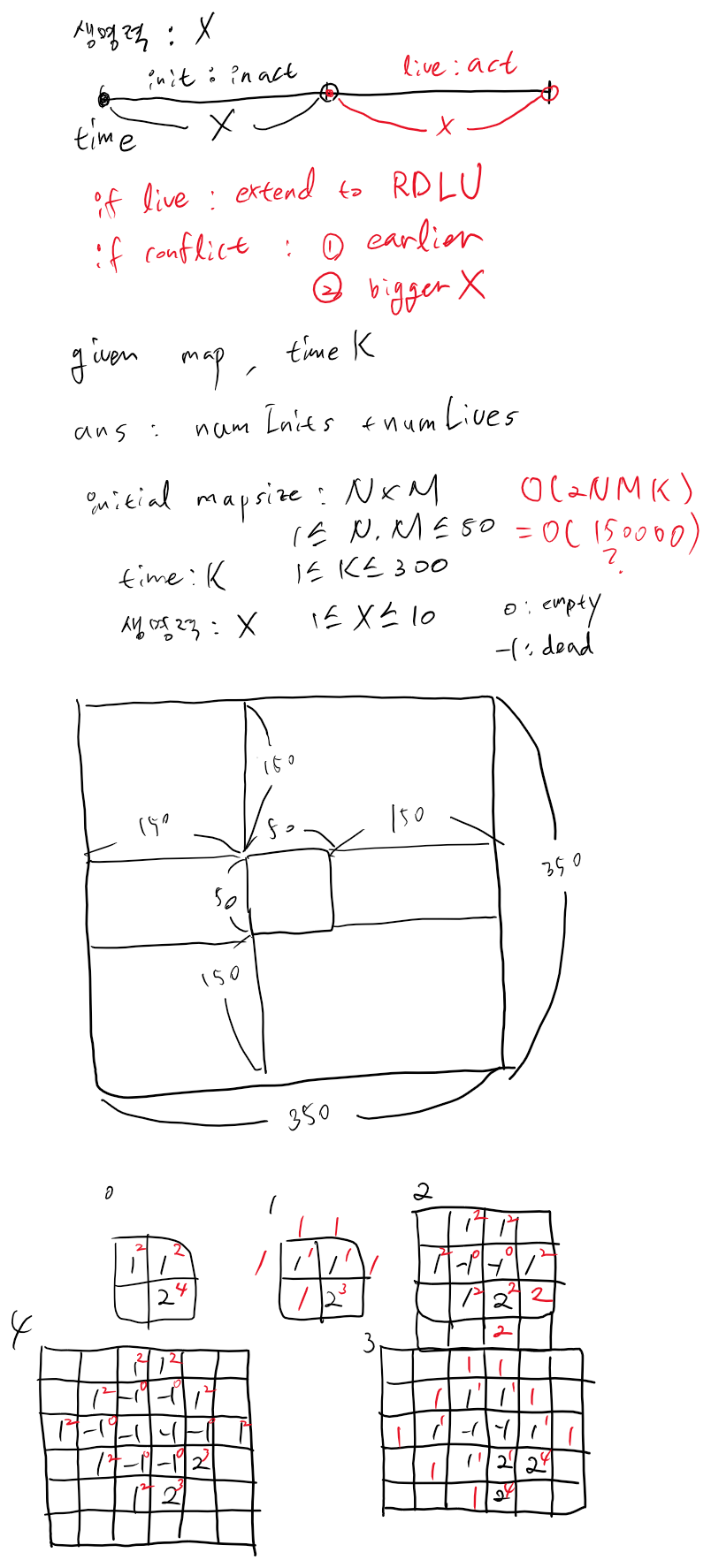
https://www.swexpertacademy.com/main/code/problem/problemDetail.do?contestProbId=AWXRJ8EKe48DFAUo
Source
explicit CProbSolve(const int N, const int M, const int K) {
(void)memset(&(g_arMap[0][0]), 0, sizeof(g_arMap));
m_initRowsN = N;
m_initColsM = M;
m_timeLimitK = K;
m_mapMaxRows = INIT_MAP_OFFSET + m_initRowsN + INIT_MAP_OFFSET;
m_mapMaxCols = INIT_MAP_OFFSET + m_initColsM + INIT_MAP_OFFSET;
m_mapRowRange.first = m_mapMaxRows;
m_mapRowRange.second = 0;
m_mapColRange.first = m_mapMaxCols;
m_mapColRange.second = 0;
FOR(row, m_initRowsN) {
FOR(col, m_initColsM) {
int val = 0;
cin >> val;
if (val > 0) {
g_arMap[0][row + INIT_MAP_OFFSET][col + INIT_MAP_OFFSET] = val;
g_arMap[1][row + INIT_MAP_OFFSET][col + INIT_MAP_OFFSET] = val;
_UpdateRange(row + INIT_MAP_OFFSET, col + INIT_MAP_OFFSET);
}
}
}
_Solve();
}
void _BFSwithGenerations() {
queue<i_ii> arqLifePos[2];
FOR_INC(row, m_mapRowRange.first, m_mapRowRange.second + 1) {
FOR_INC(col, m_mapColRange.first, m_mapColRange.second + 1) {
const int val = g_arMap[0][row][col];
if (val > 0) {
arqLifePos[0].push(i_ii(2 * val, (ii(row, col))));
}
}
}
// repeat the loop until time limit
vector<i_ii> vPrevExtendedCells;
FOR(gen, m_timeLimitK) {
const int i = gen % 2;
const int ni = (gen + 1) % 2;
if (!vPrevExtendedCells.empty()) {
// update the map and the queue with previously extended cells
_UpdateMapAndQueue(vPrevExtendedCells, arqLifePos[i]);
}
// visit all the stem cells in the current queue
while (!arqLifePos[i].empty()) {
const i_ii life_pos = arqLifePos[i].front(); arqLifePos[i].pop();
const int row = life_pos.second.first;
const int col = life_pos.second.second;
// getting old
const int life = life_pos.first - 1;
if (life <= 0) {
// dies
g_arMap[0][row][col] = -1;
continue;
}
// still alive: push into the next queue
arqLifePos[ni].push(i_ii(life, ii(row, col)));
const int val = g_arMap[0][row][col];
if (life > val) continue;
// now active!
// for extention, the map is going to be updated in the next loop
FOR(dir, eDIR_LEN) {
const int nextRow = row + DIR[dir][0];
const int nextCol = col + DIR[dir][1];
P_IFNOT(!OOR(nextRow, 0, m_mapMaxRows - 1), nextRow);
P_IFNOT(!OOR(nextCol, 0, m_mapMaxCols - 1), nextCol);
if (g_arMap[0][nextRow][nextCol] != 0) continue;
// previously empty cell
if (val > g_arMap[1][nextRow][nextCol]) {
g_arMap[1][nextRow][nextCol] = val;
_UpdateRange(nextRow, nextCol);
// extention candidates which will get old from the next after the next generation
vPrevExtendedCells.push_back(i_ii((2 * val) + 1, ii(nextRow, nextCol)));
}
}
} // while (!m_arqLifePos[i].empty())
} // FOR(gen, m_timeLimitK)
}
void _UpdateMapAndQueue(vector<i_ii> &vPrevExtendedCellsOut, queue<i_ii> &qLifePosOut)
{
while (!vPrevExtendedCellsOut.empty()) {
const i_ii life_pos = vPrevExtendedCellsOut.back(); vPrevExtendedCellsOut.pop_back();
const int row = life_pos.second.first;
const int col = life_pos.second.second;
const int val = g_arMap[1][row][col];
if (life_pos.first == (2 * val) + 1) {
// resultant extentions
g_arMap[0][row][col] = val;
// it start to get old from the next generation
qLifePosOut.push(life_pos);
}
}
}Source with respective queues for each life time
#pragma GCC optimize("O3")
#include <iostream>
#include <algorithm>
#include <memory.h>
using namespace std;
#define BASE 151
struct p {
int r, x, y;
};
int map[360][360];
p q[11][2][2001]; // life time, switching by 2, cell of each life time
int main() {
ios::sync_with_stdio(false); cin.tie(0);
register int t, tc, N, M, K, s, dx[] = { 0,0,1,-1 }, dy[] = { 1,-1,0,0 }, idx[11][2] = { 0, };
cin >> t;
for (tc = 1; tc <= t; ++tc) {
register int i, j, k, l, nx, ny, ans, siz, ni, tt;
memset(map, 0, sizeof(map));
memset(idx, 0, sizeof(idx));
cin >> N >> M >> K;
ans = 0;
for (i = 0; i < N; ++i) for (j = 0; j < M; ++j) {
nx = i + BASE; ny = j + BASE;
cin >> tt;
if (tt)
q[tt][0][idx[tt][0]++] = { 2 * tt, nx, ny };
map[nx][ny] = tt;
}
s = 0;
// for generations
for (i = 0; i <= K; ++i) {
// for each life time
for (j = 10; j >= 1; --j) {
siz = idx[j][i % 2];
ni = (i + 1) % 2;
idx[j][ni] = 0;
// for each cell of each life time
for (k = 0; k < siz; ++k) {
p &x = q[j][i%2][k];
if (x.r > j) {
if (map[x.x][x.y] > 0) {
ans++;
map[x.x][x.y] *= -1;
}
q[j][ni][idx[j][ni]++] = { x.r - 1,x.x,x.y };
}
else if (x.r == j) {
q[j][ni][idx[j][ni]++] = { x.r - 1,x.x,x.y };
for (l = 0; l < 4; ++l) {
nx = x.x + dx[l];
ny = x.y + dy[l];
if (map[nx][ny]) continue;
map[nx][ny] = j;
q[j][ni][idx[j][ni]++] = { 2 * j,nx,ny };
}
}
else if (j > x.r && x.r)
q[j][ni][idx[j][ni]++] = { x.r - 1,x.x,x.y };
else
ans--;
}
}
}
cout << '#' << tc << ' ' << ans << '\n';
}
return 0;
}GitHub